I am interested in building mathematical models to explain biological phenomena with the aim of designing better theranostic devices. Within this broad scope, I have focused on studying organelle interaction networks during my Masters and cell dynamics underlying tissue formation as an undergraduate. I find it particularly rewarding to work on problems that have a potential for translational success
So far, I have worked/am working on the following subjects:
- Exploring multiple facets of Optical coherence Tomography techniques
- Computational and Experimental analysis of Organelle Networks at the Whole Cell scale
- Evaluating the efficiency of a colonoscopy procedure
- Computational Modeling of Cellular Dynamics
- AFM observation of spatiotemporal control of Protein-DNA interaction
- Alteration of pH sensitive regime of DNA I-switch
- pH Mediated DNA Hopping
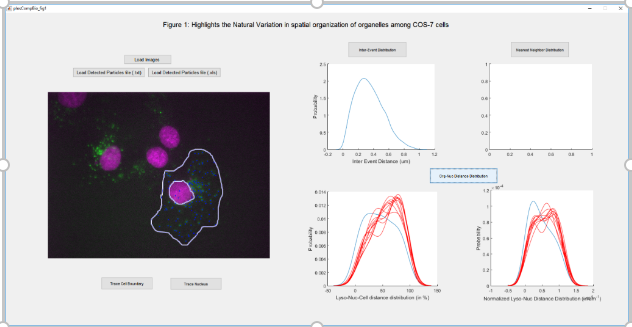 I'm fascinated by the consistency of spatial organization of organelles exhibited by a eukaryotic cell across cell-types.
Along with studying single organelle networks in isolation, the interaction amongst organelle networks interests me. To
study these in more detail, I've worked on developing spatial statistics metrics to quantify spatial distribution of homotypic organelles and
heterotypic organelle contact sites. I've also performed wet-lab experiments to actively control their spatial distribution
in order to assess the existence of homotypic/heterotypic interactions.
I'm fascinated by the consistency of spatial organization of organelles exhibited by a eukaryotic cell across cell-types.
Along with studying single organelle networks in isolation, the interaction amongst organelle networks interests me. To
study these in more detail, I've worked on developing spatial statistics metrics to quantify spatial distribution of homotypic organelles and
heterotypic organelle contact sites. I've also performed wet-lab experiments to actively control their spatial distribution
in order to assess the existence of homotypic/heterotypic interactions.
Advisor: Dr. Ge Yang
Institution: Carnegie Mellon University, Pittsburgh
Relevant Reports: Masters Thesis
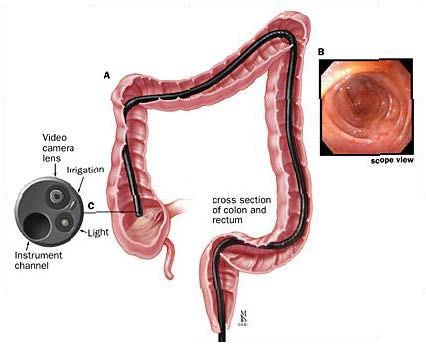 I worked with a team on designing algorithms to detect the cecum (end of colon) from colonoscopy videos and tested the
accuracy and robustness of algorithms developed to quantify distance traversed within a colon by manually snipping
videos of pre-defined length (eg. 5cm, 10cm) traversed within the colon.
The best segment of this practicum project was the opportunity to shadow Dr. Shyam Thakker (Endo/Colonoscopist) in most of
his endoscopy and colonoscopy procedures performed at West Penn Hospital, Pittsburgh.
I worked with a team on designing algorithms to detect the cecum (end of colon) from colonoscopy videos and tested the
accuracy and robustness of algorithms developed to quantify distance traversed within a colon by manually snipping
videos of pre-defined length (eg. 5cm, 10cm) traversed within the colon.
The best segment of this practicum project was the opportunity to shadow Dr. Shyam Thakker (Endo/Colonoscopist) in most of
his endoscopy and colonoscopy procedures performed at West Penn Hospital, Pittsburgh.
Advisor: Dr. Shyam Thakker, MD & Dr. Yang Cai, PhD
Institution: Allegheny General Hospital, Pittsburgh & Carnegie Mellon University, Pittsburgh
Relevant Reports: Practicum Report
 As a part of my Undergraduate Thesis, I developed an in-silico model to understand cell-population dynamics with & without the diffusional
limitation of essential nutrients. This computational tissue set-up can be used as a model system to analyse the efficacy of specific
drugs on the wound-healing process.
As a part of my Undergraduate Thesis, I developed an in-silico model to understand cell-population dynamics with & without the diffusional
limitation of essential nutrients. This computational tissue set-up can be used as a model system to analyse the efficacy of specific
drugs on the wound-healing process.
Advisor: Dr. Vignesh Muthuvijayan
Institution: Indian Institute of Technology, Madras
Relevant Reports: BTP Thesis
 I worked on Spatio-temporally controlling DNA-protein interactions at the nano-scale using visible light, and
visualized the same using an Atomic Force Microscopy (AFM) and High-Speed AFM. Apart from experimentation,
I computationally modelled the hybrid protein VVD-GAL4 to study the dynamics of DNA-protein
complex formation.
I worked on Spatio-temporally controlling DNA-protein interactions at the nano-scale using visible light, and
visualized the same using an Atomic Force Microscopy (AFM) and High-Speed AFM. Apart from experimentation,
I computationally modelled the hybrid protein VVD-GAL4 to study the dynamics of DNA-protein
complex formation.
Advisor: Dr. Masayuki Endo, Dr. Hiroshi Sugiyama
Institution: ICeMs, Kyoto University
Relevant Reports: Manuscript (in process)
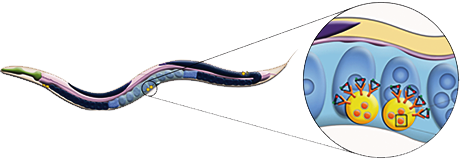 In order to study intracellular pH changes, "smart" nano-probes are required, that can simultaneously interface
with the biological system (cell) as well as relay information. In order to do so, DNA-based pH sensors were developed
at Dr. Yamuna's lab. As a summer intern, I worked on quantifying the dynamic range of modified LNA/DNA based pH sensors via FRET measurements.
Apart from in-vitro work, I was trained to work with C.elegans; from learning how to pick them at different stages of growth (L1, L2, L3, L4 and Adult) to setting up crosses.
In order to study intracellular pH changes, "smart" nano-probes are required, that can simultaneously interface
with the biological system (cell) as well as relay information. In order to do so, DNA-based pH sensors were developed
at Dr. Yamuna's lab. As a summer intern, I worked on quantifying the dynamic range of modified LNA/DNA based pH sensors via FRET measurements.
Apart from in-vitro work, I was trained to work with C.elegans; from learning how to pick them at different stages of growth (L1, L2, L3, L4 and Adult) to setting up crosses.
Advisor: Dr. Yamuna Krishnan
Institution: National Center for Biological Sciences, Bangalore
Relevant Reports: Internship Report
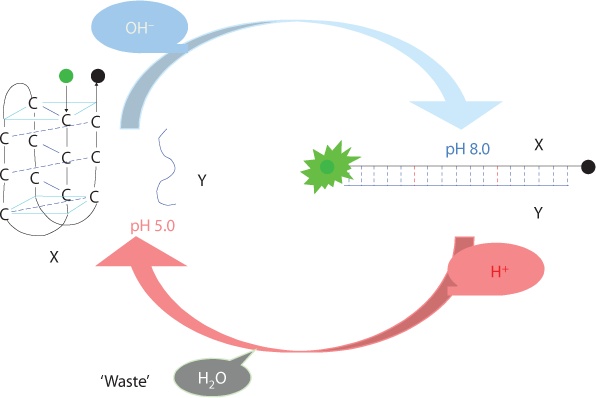 As the leader of the undergraduate team representing IIT Madras at BIOMOD (Biomolecular Design Competition) conducted by
Harvard University, we designed a Molecular Robot responsive to pH and verified the formation of the structure in-silico using NUPACK (online software) and Native PAGE (for DNA).
As the leader of the undergraduate team representing IIT Madras at BIOMOD (Biomolecular Design Competition) conducted by
Harvard University, we designed a Molecular Robot responsive to pH and verified the formation of the structure in-silico using NUPACK (online software) and Native PAGE (for DNA).
Advisor: Dr. N Manoj
Institution: Indian Institute of Technology, Madras
Relevant links: IIT Madras - BIOMOD Team